Profiling circulating tumour cells and other biomarkers of invasive cancers
Mahla Poudineh1, Edward H. Sargent, Klaus Pantel and Shana O. Kelley
During cancer progression, many tumours shed circulating tumour cells (CTCs) and other biomarkers into the bloodstream. The analysis of CTCs offers the prospect of collecting a liquid biopsy from a patient’s blood to predict and monitor therapeutic responses and tumour recurrence. In this Review, we discuss progress towards the isolation and recovery of bulk CTCs from whole blood samples for the identification of cells with high metastatic potential. We provide an overview of the techniques that initially pointed to the clinical significance of CTCs and describe the key requirements for clinical applications of CTC analysis. We also summarize recent advances that permit the functional and biochemical phenotypes of CTCs to be characterized, and discuss the potential roles of these CTC characteristics in the formation of metastatic lesions. Moreover, we discuss the use of circulating tumour DNA and exosomes as markers for early cancer diagnosis and for the monitoring of cancer progression. Next-generation technologies and biomarkers for invasive cancers should allow for the unequivocal determination of the meta-static potential of CTCs, and for the meaningful analysis of circulating tumour DNA and exosomes.
Circulating tumour cells (CTCs) and other cancer-related biomarkers are present in the blood of many patients with cancer. CTCs are believed to be involved in the formation of metastatic tumours. Indeed, high CTC levels in the bloodstream are associated with poor prognosis and an increased probability of metastatic disease1–4. It was initially thought that the formation of metastatic lesions occurs in the later stages of cancer progression. However, recent studies have shown that CTCs can leave primary tumours and enter into the circulation at a relatively early stage of tumour growth5,6. This can lead to the parallel development of met-astatic lesions and primary tumours. The study of CTCs is there-fore central to the study of the mechanism of cancer metastasis, and the analysis of CTCs and other circulating biomarkers in clinical specimens provides a basis for the development of non-invasive liquid biopsies.
We kindly thank the researchers at nature biomedical engineering for this collaboration, and for sharing the results obtained with their system.

The evolving understanding of CTC
Many studies have evaluated how CTCs can be used as a predic-tor of clinical outcome. The CellSearch system — a device for CTC analysis cleared by the US Food and Drug Administration (FDA) — enriches and enumerates CTCs from peripheral blood, and has been used widely for CTC enumeration in the clinic. It counts epithelial cancer cells from whole blood using magnetic immunotargeting of the epithelial cell adhesion molecule (EpCAM), and subsequently identifies CTCs with fluorescently labelled antibodies against cyto-keratin7. The presence of CTCs detected using CellSearch is associ-ated with poor prognosis in metastatic and localized carcinomas, and the significance of CellSearch results has been demonstrated in a series of prospective clinical trials. A cohort study that evalu-ated the prognostic role of CTC counts in localized and metastatic colorectal cancer patients showed that the presence of CTCs is asso-ciated with reduced survival in colorectal cancer patients8. A multi-centre study of metastatic breast cancer patients validated the utility of the CellSearch system to detect CTCs in the blood of patients7.Other studies have shown that low levels of CTCs before and dur-ing chemotherapy were associated with a better clinical response in metastatic breast cancer9. Analysis of CTC count using CellSearch in patients with castration-resistant prostate cancer showed that decreases in CTC count after treatment correlated with improved overall survival following abiraterone treatment and chemother-apy10. Several clinical studies also tracked the presence of CTCs before and after surgery for non-metastatic breast cancer11, colorec-tal cancer12 and bladder cancer13. A recent study of non-metastatic breast cancer patients receiving neoadjuvant chemotherapy demon-strated that the CTC count was an independent prognostic factor from the pathological response of the primary tumour, thus indicat-ing that systemic therapy can have differential effects on primary and disseminating tumour cells14.
Although it appears that CTC counts are good predictors of prognosis and of the efficacy of therapy, it has become clear that simple enumeration of CTCs is not sufficient. Increasingly, more insight is available pertaining to the physiology of metastasis, and to the involvement of several stages in the metastatic cascade, includ-ing trans-endothelial migration and intravasation of tumour cells into the circulation, cell survival in the circulation, transport of cells through the vasculature followed by extravasation, and coloniza-tion and formation of metastatic lesions (Fig. 1)15,16. It is clear that patients have CTCs of varying phenotypes, and that only a small fraction of the CTCs in circulation have the potential to participate in all of these events. Most cancer cells are rapidly destroyed in the circulation by the immune system or by haemodynamic forces17, which may be a frequently underestimated rate-limiting step in metastasis that could explain why the detection of intact CTCs is closely correlated to tumour recurrence. The ability of cells to extravasate into the surrounding tissue by degrading the extracel-lular matrix is another rate-limiting step in metastasis, making it a highly inefficient process. Experimental studies have shown that only a minority of the CTCs that survive form macrometastases18–20,
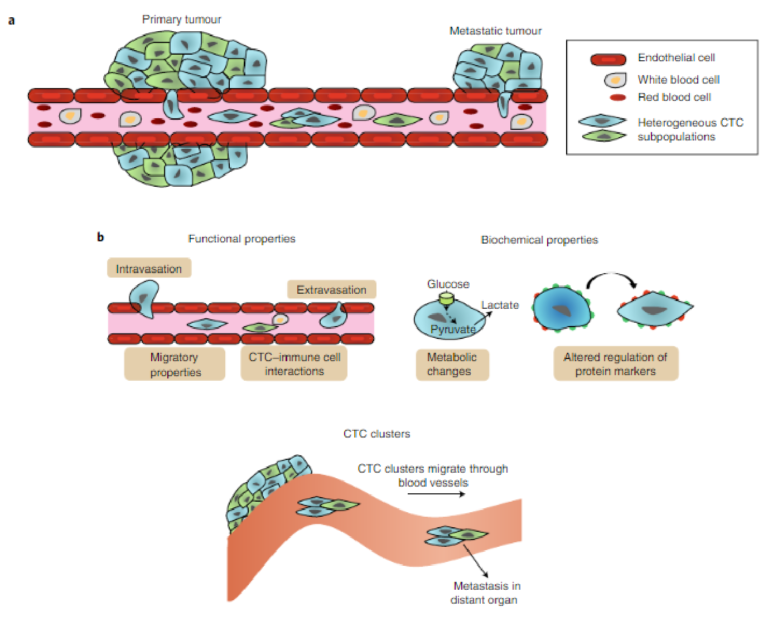
Fig. 1 | CTC phenotypic properties. a, The characterization of the phenotypic properties of CTCs is critical for evaluating their metastatic potential. Heterogeneous CTCs enter the bloodstream and migrate through blood vessels. Further dynamic heterogeneity may arise as the cells circulate. b, Functional and biochemical phenotyping may enable the evaluation of the invasiveness of CTCs. By measuring migration potential and cell–cell interactions, the likelihood of intravasation and extravasation can be gauged. Tracking biochemical properties is helpful to define CTC subpopulations and study the dynamics of heterogeneity. Monitoring CTC clusters is important, because aggregates of cells appear to have greater metastatic potential.
thus confirming the inefficiency of metastatic colonization. However, autopsy studies have shown that cancer patients who had only a few detectable overt metastatic lesions can harbour hundreds of small, occult metastases. Thus, the exact degree of metastatic inefficiency in cancer patients remains unknown.
The dissemination of CTCs from primary tumours can occur through phenotypic changes that are accompanied by the epithelial to mesenchymal transition (EMT), a process that may therefore play an essential role in metastasis4,21. The EMT is characterized by the downregulation of epithelial markers such as EpCAM, cytokeratins and E-cadherin, and by the upregulation of mesenchymal markers such as vimentin and N-cadherin. Interestingly, studies on thou-sands of cancer patients have shown that CTCs expressing epithe-lial markers such as cytokeratins can be frequently detected, which suggests that the EMT for tumour-cell release into the blood22 is not absolutely required. Nevertheless, it cannot be excluded that these CTCs have undergone a partial EMT. The loss of the epithelial characteristics that ensues during the EMT engenders CTCs with greater motility and, more broadly, CTCs with characteristics that support the invasion of secondary sites23,24. Studies of samples col-lected from human cancer patients have shown that the EMT is a dynamic process, and that progression towards a mesenchymal phenotype is associated with poor outcomes25. Nevertheless, the number of studies demonstrating the prognostic relevance of cyto-keratin-positive CTCs is by far greater22. One study classified CTCs from patients with liver, nasopharyngeal, breast, colon, gastric and non-small cell lung cancers on the basis of EMT markers. CTCs were detected in patient samples, and three CTC subpopulations were identified using EMT markers — epithelial CTCs, epithelial and mesenchymal CTCs, and mesenchymal CTCs26. However, it should be noted that experimental studies have shown that tumour cells that underwent the complete EMT process are unable to form metastases, whereas that cells with an intermediate phenotype have the highest plasticity to disseminate and form secondary lesions in distant organs22. During cancer progression, the phenotypic plas-ticity induced in CTCs by the EMT is revealed through the occur-rence of a reverse process, the mesenchymal to epithelial transition (MET), which involves the conversion of cells in a mesenchymal state into epithelial derivatives. Compared with the EMT, the MET is poorly understood. It is believed that by allowing cancerous cells to regain epithelial properties, the MET participates in the establish-ment and stabilization of distant metastases24,27.
Given the importance of the phenotypic properties of CTCs and the influence of these properties on the prolonged survival of CTCs in the bloodstream and on their capacity to form metastatic tumours, advanced methods that allow for more detailed charac-terization of CTCs are critically needed. Also, many questions con-cerning how the release of CTCs coincides with disease progression, differences between CTCs generated by primary tumours and those generated by metastatic tumours, and the role of the EMT in metas-tasis, will require detailed studies of these cells in cancer models and in patients.
Recent advances in technologies for CTC capture
CTCs can provide a wealth of information that reveal a tumour’s molecular profile and facilitate the dynamic monitoring of cancer progression. The information collected from these cancer biomark-ers can be used to classify and inform the treatment of patients. In light of the significance of CTCs in cancer development and metas-tasis, significant effort has been directed towards the development of advanced methods for the capture and characterization of these cancer biomarkers. Although CTCs were first observed in a patient during autopsy in 1869 (ref. 28), techniques for the isolation of these cells were only first reported around 1960 (refs 29,30). The field started to advance rapidly when immunomagnetic separation allowed the highly specific separation of CTCs in the late 1990s (ref. 31), but the recognition that CTCs can be highly heterogeneous32,33 high-lighted the need to separate subpopulations and to develop more marker-agnostic separation technologies (Fig. 2). The development of microfluidic systems for CTC isolation and analysis over the past decade has increased isolation efficiencies signficantly34–37, and has spurred accelerated progress in this research area. In 2016, the first CTC-based test was validated clinically for therapeutic selection in prostate cancer, indicating that the progress made in CTC research was also having significant clinical impact38,39.
Technologies for CTC capture have been reviewed23,40–42, and therefore below we only highlight the most advanced approaches, with an emphasis on technologies that are now commercially avail-able. Although many new technologies have in recent years been developed for the detection and molecular profiling of CTCs, the majority have yet to demonstrate clinical utility. At present, the CellSearch system is the only FDA-cleared method for CTC detec-tion for clinical applications. Yet new tests that detect important drug-resistance markers in CTCs are emerging38. Stringent clinical validation of new devices is required before their introduction to the management of cancer patients, and even the early validation of new technologies should be performed with clinical specimens to ensure that the techniques have sufficient levels of sensitivity and specificity.
A key technical challenge related to the analysis of CTCs is their extreme rarity in the circulation (relative to normal blood cells), which makes their capture challenging6. Recent advances in rare-cell capture technology enable the isolation of CTCs with increasingly high sensitivity and specificity34–36,43–49. Several of the first assays to become commercially available isolated CTCs using magnetic immunoaffinity labelling50–52. Beads coated with antibodies spe-cific for CTC surface markers were used to target tumour cells in blood (Fig. 3a), with an external magnetic field used to isolate CTCs for further analysis. Most of the techniques reported to date use EpCAM as the target marker to enumerate the bulk population of CTCs. EpCAM is a specific cancer-cell surface marker that is not expressed on normal blood cells and that has levels that are known to vary during cancer progression53,54. CTC subpopulations with high levels of EpCAM expression are isolated, but CTCs with low or even no EpCAM expression can be lost during the capture pro-cess55–57. Although conflicting results have been reported concern-ing whether EpCAM-negative cells are clinically important58–60, it nevertheless seems to be favourable to capture all subsets of CTCs for further evaluations. Hence, using cocktails of antibodies for CTC capture is increasingly popular36,61.
The use of physical parameters for cell separation has also been applied successfully to the isolation of CTCs. A size-based separa-tion method that uses a high-throughput vortex chip (Vortex HT) has facilitated the label-free isolation of rare tumour cells (Fig. 3b)62. The Vortex HT chip consists of rectangular reservoirs, which gener-ate laminar fluid microvortices at high flow rates to passively purify large CTCs from whole blood samples. The Parsortix device (Angle Plc) also enables automated size-based selection, and has been applied to the isolation of CTCs in a variety of cancer types63. The AccuCyte–CyteFinder assay (Rarecyte) isolates CTCs on the basis of cell density, followed by the identification, analysis and retrieval of rare CTCs. The assay allows for the sensitive identification of rare cells and for the interrogation of relevant disease biomarkers64.
Dielectrophoresis (DEP), which relies on electrokinetics, can also be used for tumour-cell isolation. DEP depends on the cells’ dielectric properties, which reflect cell morphology and membrane surface area. Under a non-uniform electric field provided by an array of electrodes, cells with different dielectric properties display differential transport rates. DEP-based isolation can have a relatively high output purity, owing to the specificity of the dielectric pheno-types of different cell types65–67. In the DEPArray system (Fig. 3c),
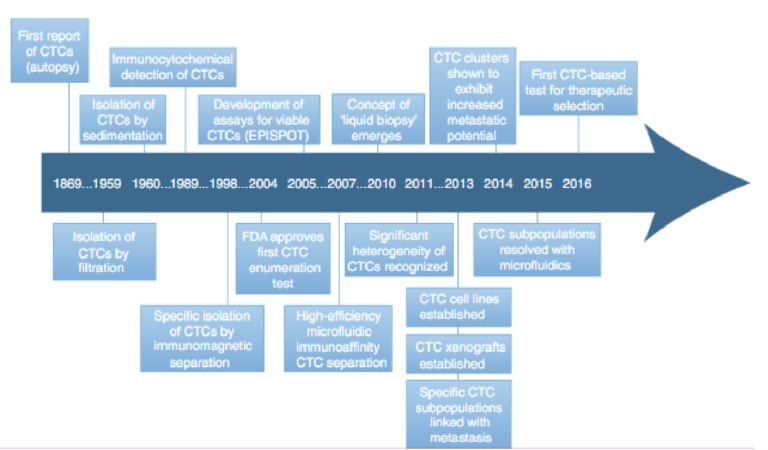
Fig. 2 | Timeline of significant discoveries related to CTC biology, to the clinical significance of CTCs and to technologies that have advanced the understanding of the properties of CTCs and their clinical utility. The development of microfluidic devices has significantly accelerated the pace of CTC research and of the clinical translation of CTCs over the past decade
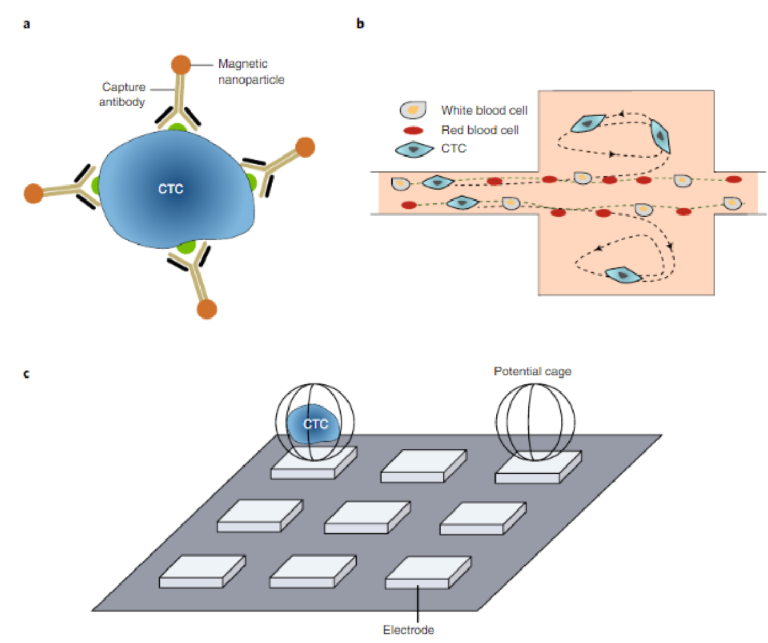
Fig. 3 | Rare-cell capture technologies isolate CTCs with high levels of sensitivity and specificity. a, Immunomagnetic isolation. Many immunoaffinity capture assays, including that carried out by the CellSearch system, label CTCs with magnetic particles functionalized with an antibody against a specific surface marker. b, Size-based separation. At high flow rates, the Vortex chip generates microvortices that allow cells larger than a threshold size to be trapped while smaller cells (red and white blood cells) pass through62. c, DEP-based single-cell capture. In the DEPArray system, electric fields generate potential cages that trap single cells69. Panel b adapted from ref. 62 under a Creative Commons licence CC BY 3.0. Panel c reproduced from ref. 69, IEEE
commercialized by Silicon Biosystems, single cells are deposited into individual DEP cages generated with electric fields68,69. This system is a particularly appropriate approach for the isolation of CTCs for subsequent offline characterization.
An alternative approach, commercialized by Epic Science, analy-ses patient blood samples without CTC enrichment or depletion of leukocytes. After the red blood cells have been removed, the remaining cells are dispersed on a microscope slide and are stained for cancer markers. Image analysis is performed, and the stained cells are analysed on the basis of morphology. Similar to other CTC assays, the Epic assay is also integrated with downstream capabilities for the evaluation of protein biomarkers (via immunofluorescence) and genetic biomarkers (through fluorescence in situ hybridization or next-generation sequencing)70,71. Side-by-side comparisons with other CTC technologies in independent laboratories and clinical validation of the Epic assay are ongoing.
These capture devices isolate the bulk population of CTCs. Yet it is imperative to differentiate subpopulations that may have varying phenotypes with different levels of clinical relevance. Several stud-ies have shown that distinct CTC subpopulations can have differ-ing levels of metastatic potential59,60. One of these studies59 collected large patient samples and separated the CTCs into subpopulations bearing different surface markers. The different subpopulations were then implanted in animals to establish which CTCs promoted the formation of tumours. This type of approach is an excellent means to establish signatures for invasive CTCs; yet it is challeng-ing to scale and implement routinely because hundreds of CTCs are required, which are rarely found in the volume of blood typi-cally collected from cancer patients. Advanced rare-cell-profiling tools72,73 enable fingerprinting of genomic and proteomic proper-ties; however, even the most advanced techniques reported to date have performed these analyses offline (and require extensive cel-lular manipulation that may influence the characterization results). New techniques that integrate CTC capture with characterization technologies to ensure that accurate information can be collected about their status are thus needed. To be effective, such technologies would require the following characteristics
:• Characterization technologies should enable the isolation of CTCs with a high level of specicity
.• Capture methods should handle low numbers of CTCs (less than 100 target cells per 109 blood cells)
.• Analysis methods should exhibit high levels of performance with whole blood samples, and be validated with patient samples
.• Profling approaches should enable the sorting of heterogeneous CTC subpopulations to distinguish cancer cells with high and low metastatic potential
.• Capture techniques should allow the release of viable cells to enable downstream analyses.
The increasing availability of methods and devices that enable high-efficiency CTC capture will continue to facilitate the exploration of the clinical relevance of these cells and of their significance for the biology of disease. However, moving forward, it will be essential that information from functional profiling is collected during the capture of CTCs from clinical specimens.
Monitoring the functional phenotypes of CTCs
The ability to characterize the functional traits of CTCs, such as can-cer-cell migration, is advancing rapidly. CTC migration is involved in all steps of tumour-cell dissemination and therefore is an impor-tant functional property to assess74–76. CTCs enter into the blood cir-culation and migrate through the invasion of surrounding tissue or the stimulation of external forces. At present, two mechanisms are thought to be involved in the detachment of CTCs during tumour growth: a passive mechanism, whereby CTCs are mechanically trapped within the microvasculature and migrate under the influence of external stimulators such as mechanical or chemical forces; and an active mechanism, whereby tumour cells intrinsically gain the ability to migrate by modifying their cell morphology, position and surrounding tissue5. Cancer cells may migrate as single entities or as a cluster of cells. Collective cell migration requires strong cell–cell adhesion. However, single cells may infiltrate when they lose the adhesive bonds with neighbouring tumour cells.
The development of microfluidic approaches has enabled the study of the migration of tumour cells with single-cell resolution. Recent results suggest that the collective or individual migration of tumour cells corresponds to their relative expression of epithelial and mesenchymal biomarkers. This idea was confirmed by studying the migration of tumour cells using an enclosed array of polydimethylsiloxane (PDMS) micropillars that periodically disrupted cell–cell contacts and enhanced individual scattering77. Cells surrounded by many neighbours migrated collectively and showed high levels of epithelial markers. However, cells expressing high levels of mesen-chymal markers tended to move with few nearest neighbours, and dispersed efficiently with fast and straight trajectories (Fig. 4a). In another study, migration patterns and velocities of single cells were monitored by using an array of miniaturized chambers (Fig. 4b)78. EMT-induced cells showed more aggressive migration phenotypes, and the highest velocities were observed for cells that exhibited sig-nificant levels of drug resistance.
Tumour cells can move both randomly and directionally; how-ever, invasion, migration and dissemination are most efficient when cells are involved in directed migration caused by external stimulators79. Growth factors and chemokines mediate CTC migration through chemotactic migration. One approach that investi-gated the effects of a chemical gradient on cell migration used a method with single-cell resolution that enables the post-migration collection and analysis of cell subpopulations with different chemotactic behaviours (Fig. 4c)80. A variety of aspects of cancer-cell migration has been explored using microfluidic devices; how-ever, many studies were confined to cultured cancer cells rather than CTCs obtained from blood samples. A recently reported device allowed the quantitative study of single-cell migration for actual CTCs obtained from xenograft cancer models, and showed that cancer-cell subpopulations with different levels of epithelial marker expression exhibit varying chemotactic migration profiles (Fig. 4d)81. The xenograft models of prostate-cancer cell lines showed that CTCs extracted from animals with less aggressive tumours exhibited insignificant levels of chemotaxis; in contrast, for the animals bearing aggressive tumours, the majority of CTCs displayed high levels of migratory behaviour. This study provided direct evidence that phenotypic subpopulations of CTCs may exhibit differing levels of invasiveness.
The ability of CTCs to enter into the bloodstream (intravasation) and the process by which cancer cells transmigrate across a monolayer into a model extracellular space (extravasation) are two functional phenotypes of tumour cells that play important roles in metastasis and cancer-cell dissemination82,83. Microfluidic devices have helped elucidate that tumour related biochemical factors and other cells present in the tumour microenvironment (such as macrophages) control the ability of tumour cells to enter the bloodstream84,85. A microfluidic-based assay developed to model the tumour/vascular interface enabled quantification of endothe-lial barrier function, and showed that the tumour cells invade in response to the externally applied growth-factor gradients or to cell-to-cell communication85. The assay also allowed for the monitoring of the role of biochemical factors and of cellular interactions in the regulation of cancer-cell intravasation. An approach to model cancer-cell extravasation was developed using a microfluidic system consisting of three media channels that are separated by a collagen-gel matrix86. The results showed that extravasation events occur within the matrix in the first 24 hours following the introduction of the cancer cells, and that the events are associated with a significant increase in the permeability of the endothelial monolayer. Although these model systems have facilitated some of the first studies of these phenomena, they have not yet been validated using patient CTCs, and the underlying mechanisms of intravasation and extravasation remain poorly defined.
Studying the migratory behaviour of CTCs presents a means of identifying cells with greater invasive capacity. Many factors play roles in cancer-cell migration and in their ability to interact with other cells. Thus, microfluidic design principles that integrate many of these factors can contribute to the development of more sophisticated models of tumour cell dissemination.
Tracking the biochemical properties of CTCs
Next-generation sequencing has been used to characterize CTCs, and has identified discordance between the gene expression lev-els of CTCs and of cancer cells from their corresponding primary tumours, signifying the existence of distinct CTC subpopulations that contribute to metastasis87,88. The development of personal-ized medicine for cancer patients relies on the identification of the molecular drivers of the disease. Biomarkers predicting therapy response measured from tumour biopsy samples are not without ambiguity, because tumours continually evolve at the molecular level, and a single-site biopsy sample may not accurately represent a patient’s disease. Genetic alterations in baseline CTCs may correlate with clinical outcome, identifying CTCs as an objective biomarker to define the personalized therapy of an individual cancer patient before the start of treatment89. Molecular analysis of CTCs has been recently employed to predict whether small-cell lung cancer patients are chemosensitive or chemorefractory. In particular, copy-number aberrations in CTCs were examined from pretreatment small-cell lung cancer blood samples, and a classifier based on these aberrations was generated90. These studies highlight the importance of continual monitoring of the biochemical properties of CTCs of patients for optimal treatment efficacy.
It is crucial to detect viable CTCs in the peripheral blood of cancer patients, because only functional cells are able to contribute to the formation of metastatic lesions. An enzymatic assay, referred to as EPISPOT91,92, uses the secretion, shedding or active release of specific protein markers to distinguish viable CTCs from apoptotic CTCs (Fig. 5a). After an enrichment step, CTCs are cultured in plates coated with an antibody against a specific protein marker for 24–48 hours. During the incubation step, viable CTCs secrete pro-teins that are directly captured on an antibody-coated membrane. The protein marker is then detected using a secondary antibody. This assay can be combined with an enrichment system to detect viable CTCs. Indeed, CTCs isolated from the blood of patients with breast, prostate and colon cancers have been analysed using the EPISPOT assay. Overall survival was linked with the measured CTC status, which allowed the stratification of patients in low-risk and high-risk groups and demonstrated the clinical relevance of viable CTCs91,93,94
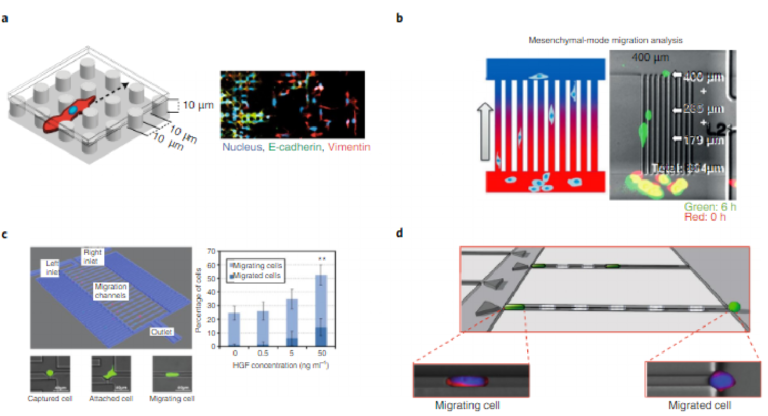
Fig. 4 | Devices for the characterization of migratory behaviour (a functional phenotype of CTCs). a, Collective and individual migration profiling. A micropillar array that facilitates migration profiling77. Cells were stained to visualize the nucleus (blue), E-cadherin (green) and vimentin (red). b, Mesenchymal-mode migration analysis. An array of channels that allows the velocity of mesenchymal cells to be monitored78. Red and green indicate cell positions at the beginning of the assay and after six hours, respectively. c, Single-cell migration characterization. A microfluidic device that permits tracking of migration at the single-cell level80. The graph illustrates the relative ratio of completely migrated cells to those that are still migrating; they were detected within the channel as a function of the chemoattractant (hepatocyte growth factor (HGF)) concentration. d, Chemotactic migration analysis. A cell-capture device that isolates cells with differing biochemical phenotypes and then allows the evaluation of chemotaxis74. Panels adapted from: a, ref. 71, Macmillan Publishers Ltd; b, ref. 78, Wiley; c, ref. 80, Macmillan Publishers Ltd; d, ref. 81, Wiley
Deconvolution of the heterogeneity of CTCs requires methods that can provide highly detailed phenotypic profiles. To meet the challenge of high-resolution profiling of CTCs, a device-based approach, magnetic ranking cytometry, was developed to generate a phenotypic profile based on surface expression at the single-cell level (Fig. 5b)45. Finely resolved sorting of CTCs was achieved via the introduction of differently sized nickel micromagnets along a microfluidic channel. The analysis of samples drawn from mice and from human cancer patients showed that the strategy enables the dynamic properties of CTCs to be tracked at the single-cell level as a function of tumour growth and aggressiveness.
Profiling of multiple proteins (including intracellular markers) in single CTCs enhances the understanding of rapidly evolving CTC biology. A recent advance in microfluidic western blotting enabled the measurement of a panel of proteins in single CTCs isolated from patient samples (Fig. 5c)95. This single-cell western blot (scWB) approach quantifies eight surface and intracellular proteins in individual CTCs by using a microfluidic-targeted proteomics tool, and allows for estimates of variations in protein expression among CTCs. The assay was able to normalize target protein expression by the number of CTCs analysed per assay in patients with metastatic breast cancer. The scWB assay can be used to integrate upstream functional screens, to quantify the cellular response to pharmaceu-tical agents and to advance the optimization of the performance of affinity reagents by facilitating library screens.
Cellular metabolism may also be perturbed in CTCs96,97. During cancer progression, tumour cells reprogramme their metabolic behaviour. Some of the most striking changes in tumour cellular bioenergetics include increases in glycolysis, elevation of glutami-nolytic flux, upregulation of amino acid and lipid metabolism, and enhancement of mitochondrial biogenesis98. Furthermore, changes in cancer-cell metabolism that result in the acidification of the extracellular environment create favourable microenvironments for the activation of proteases, including matrix metalloproteinases (MMPs). The activity of the MMP family of enzymes, specifically MMP-2 and MMP-9, plays a pivotal role in the multistep processes of invasion and metastasis, including proteolytic degradation of the extracellular matrix99. A meta-analysis showed that the overex-pression of MMP-9 and MMP-2 in serum may be correlated with poor prognoses in breast cancer100. A device composed of hydrogel microwells allowed for the capture of cancer cells and for the fluo-rescence-based detection of cell-secreted proteases such as MMP-9 (Fig. 5d)101 with high sensitivity and specificity. Using a combina-tion of hydrogel microwells and reconfigurable microfluidics, the study demonstrated detection of MMP-9 released from as few as 11 cells. In another study, an electrochemical immunosensor consist-ing of an assembly of gold nanoparticles on nitrogen-doped gra-phene sheets provided robust immobilization of antibodies suitable for the ultrasensitive detection of MMP-2 (ref. 102). The feasibility of the immunoassay was verified via the analysis of clinical samples.
Distinct molecular features of cancer metabolism can be observed through imaging modalities such as positron-emission tomography, magnetic resonance spectroscopy imaging and mag-netic resonance imaging, which play an indispensable role in clini-cal oncology103. Cancer metabolism has been extensively exploited for the initial diagnosis and staging of cancer, and for monitor-ing tumour responses to therapies and detecting tumour recur-rence. Such non-invasive diagnostic methods can accurately detect changes in biological processes within primary tumours and meta-static sites over an extended period of time (with respect to the cor-responding changes in normal surrounding tissues). Furthermore, it has become apparent that cancer metabolic processes are highly heterogeneous103. In this regard, single-cell techniques such as scWB can provide unprecedentedly detailed information.
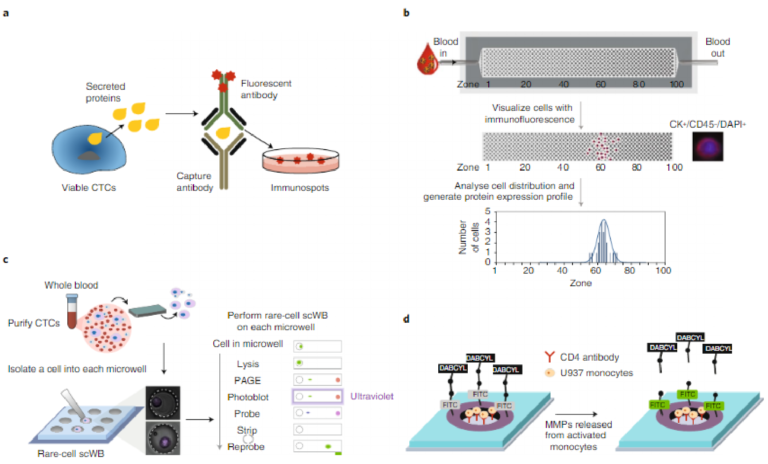
Fig. 5 | Devices and assays for the characterization of the biochemical phenotypes of CTCs. a, Secreted protein analysis. The EPISPOT assay profiles proteins secreted from live CTCs91. b, Surface protein analysis. Magnetic ranking cytometry arranges nanoparticle-labelled CTCs into one of 100 capture zones according to how many nanoparticles are bound to the cell surface. The cells are immunostained and the distribution of cells reports on surface-expression levels of a target protein as well as on the degree of cell heterogeneity45. CK+, cytokeratin; CD45–, leukocyte common antigen, a marker specific for white blood cells; DAPI, 4,6-diamidino-2-phenylindole. c, Single-cell immunoblotting. A single-cell western-blotting approach allows immunoblotting to be performed on single CTCs95. PAGE, polyacrylamide gel electrophoresis. d, Protease release analysis. A device that measures proteases secreted from cancer cells using a fluorescence readout101. The glass regions opened in the gel layer were functionalized with CD4 antibodies for cell capture. Cells bound within the hydrogel are stimulated to release MMP-9, and protease molecules then diffuse into the gel and cleave a peptide that fluoresces once the cleavage occurs (MMP-9-specific peptides were modified with a donor/acceptor fluorescence resonance energy transfer pair (fluorescein isothiocyanate (FITC) and 4-(dimethylaminoazo)benzene-4-carboxylic acid (DABCYL)) and covalently linked into the gel). Panels adapted from: b, ref. 45, Macmillan Publishers Ltd; c, ref. 95, Macmillan Publishers Ltd. Panel d reproduced from ref. 101, American Chemical Society.
Rapid progress has been made in the development of tech-niques that isolate and characterize CTC subpopulations according to biochemical phenotypes. However, devices typically monitor a single type of phenotype. There remains a need for devices that can provide comprehensive profiling104,105. For example, new microflu-idic strategies may interrogate the effect of changes in cancer-cell metabolism, as well as the effects of altered regulation of epithelial and mesenchymal markers on cell migration.
Metastasis-promoting CTC clusters
Cancer cells can also infiltrate as a cluster of cells. These clusters, which can be directly derived from primary tumours or from the aggregation of single CTCs, are present in the blood of cancer patients, and their contribution to metastasis is being intensely pur-sued106. CTC clusters appear to have increased metastatic potential when compared with single CTCs. In addition, CTC clusters with different sizes or components have distinct biological and physical characteristics that promote their metastatic potential. CTC clusters can comprise both tumour cells and non-tumour cells such as mesen-chymal cells, epithelial cells, cancer-associated fibroblasts, immune cells and platelets107,108. The presence of these non-malignant com-ponents may increase the viability of tumour cells within the clusters and contribute to metastatic efficiency. For instance, platelets in CTC clusters physically shield the tumour cells, thereby protecting them from host immune surveillance and blood-shear damage109.
Microfluidic approaches can detect CTC clusters by taking advantage of the clusters’ physical and biological properties. A microfluidic device, the Cluster-Chip, isolates CTC clusters from whole blood samples while allowing single CTCs to pass through the structures110. The device contains multiple rows of shifted tri-angular pillars forming consecutive cluster traps (Fig. 6a). The Cluster-Chip successfully trapped CTC clusters from patients with metastatic breast, prostate and melanoma cancers.
Although it was thought that CTC clusters are incapable of transiting through narrow blood vessels owing to the large size of the clusters, many reports claim to have achieved the isolation of CTC clusters. The assumption was challenged through the use of a microfluidic device mimicking capillary constrictions (Fig. 6b)111. The device showed that CTC clusters isolated from breast and mel-anoma patients rapidly and reversibly unfold to single-file chains through the cleavage of intracellular adhesions. Clusters containing up to 20 cells successfully traversed the constrictions even in whole blood. Hence, CTC clusters may contribute to tumour dissemina-tion to a larger extent than previously thought.
For a number of cancer types, the expression of cancer stem-like markers (such as CD44, ALDH7A1 and KLF4) correlates with the occurrence of metastasis and with reduced patient survival112. This suggests that cancer cells enriched in stem-like features may be the precursors of metastasis. Recent studies indicate that CTC clusters have a high degree of both molecular and cellular heterogeneity106,113, and that only a few cells within a CTC cluster retain stem-like prop-erties. In fact, the stem-like tumour cells within the CTC clusters are thought to be responsible for the formation of metastatic lesions
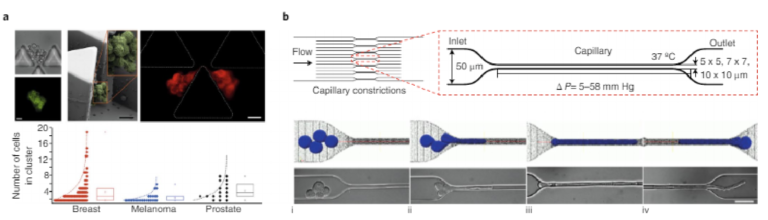
Fig. 6 | Analysis of CTC clusters. Clusters of CTCs have been shown to have increased metastatic potential and to remain intact in the bloodstream. a, Cluster capture and characterization. A device that specifically captures CTC clusters on the basis of their size110. Multiple rows of shifted triangular pillars form consecutive cluster traps. Top: images of CTC clusters: brightfield and fluorescent images of a CTC cluster stained for cancer surface markers (top left); scanning-electron-microscopy image of a fixed CTC cluster (top middle); fluorescent image of a highly deformable CTC cluster stained for cytokeratin (top right). Scale bars, 20 μ m. Bottom: size distribution of CTC clusters isolated from different cancer patients. b, Cluster migration analysis. Analysis of cluster integrity using a device that mimics small blood capillaries111. The device contains 16 parallel microchannels with square cross-sections of different sizes, to mimic capillary-flow conditions. CTC clusters travel through these constrictions at 37 °C and under physiological pressure gradients (∆ P). The micrographs (bottom) show a four-cell cluster (i) approaching, (ii) elongating, (iii) migrating and (iv) exiting the capillary constriction. Scale bar, 50 μ m. Panels adapted from: a, ref. 110, Nature America Inc.; b, ref. 111, PNAS
The study of CTC clusters may transform the understanding of cancer metastasis. However, many challenges need to be over-come before CTC-cluster analysis can be adapted for clinical use. Techniques for the specific isolation of CTC clusters that preserve their original status and that enable the in-depth study of CTC clus-ters that are physiologically relevant are needed. Further identifi-cation of stem-cell markers that robustly define cancer stem cells within a highly heterogeneous CTC cluster and that report on the cells’ status within the metastatic cascade is also needed. Moreover, the direct molecular mechanisms by which these stem-cell mark-ers contribute to the metastatic cascade will need to be investigated. And large-scale studies would need to compare the clinical rel-evance of single CTCs and CTC clusters
Strategies for the characterization of cancer biomarkers
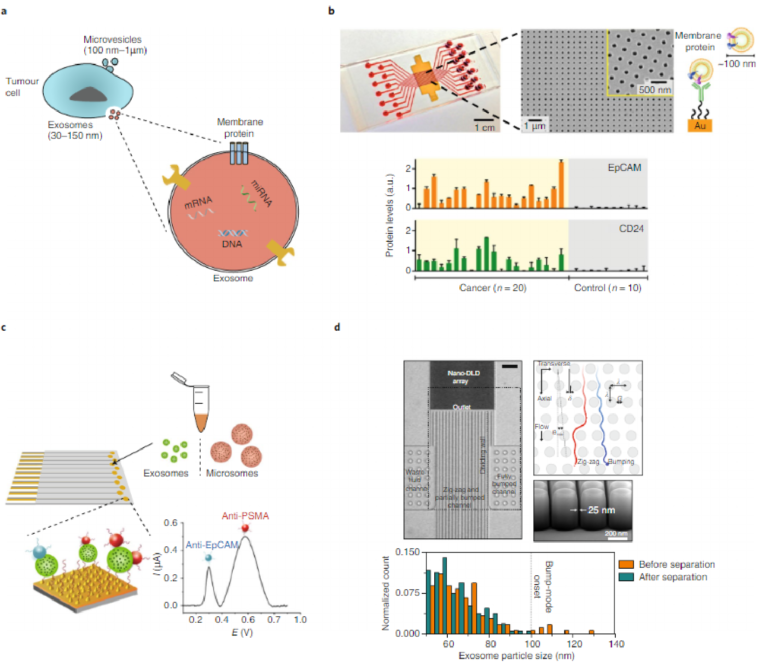
Substantial progress has been made in the discovery of new circulat-ing cancer biomarkers, including exosomes and cell-free circulating tumour DNA (ctDNA)114,115. Exosomes are small membrane-bound cell fragments, between 30 and 150 nm in diameter, and have recently emerged as a new class of cancer biomarkers116. Exosomes are actively secreted by both healthy and cancer cells. The exosomes shed by cancer cells are known to carry proteins, functional messen-ger RNAs, microRNAs and DNAs that are similar to those of their host tumours; hence, with respect to CTCs, exosomes offer signifi-cant advantages as biomarkers, including abundance, diversity and stability, which make them promising markers for cancer diagnosis and cancer monitoring. Also, exosomes exist in large quantities in accessible biofluids such as blood, urine, saliva and ascites117. They have been identified even in tumours that do not release detectable CTCs. For example, exosomes were found in samples from patients with ovarian, colorectal, brain and breast cancers118–120. Moreover, exosomes may carry the molecular markers of primary tumours including proteins and nucleic acids, and have been shown to contribute to modulating the tumour microenvironment and to directing the site-specific homing of CTCs121 (Fig. 7a). In light of the roles of exosomes in cancer progression, a variety of approaches have been developed to characterize these cancer biomarkers.
A commonly used approach for exosome isolation uses high-speed centrifugation. However, ultracentrifugation approaches are time-consuming and require expensive laboratory equipment. One solution to this problem uses affinity-based purification, which can be executed using an array of periodic nanoscopic holes that are functionalized with specific antibodies (Fig. 7b)122. This approach identified exosomes derived from ovarian cancer cells on the basis of CD24 and EpCAM expression. Another assay for exosome char-acterization employed a multiplexed electrochemical sensor that enabled the detection of exosomes via direct electro-oxidation of metal nanoparticle labels (Fig. 7c)123. Metal nanoparticles with dif-ferent oxidation potential were conjugated to recognition agents for specific protein markers expressed on exosomes. The assay enabled the direct analysis of exosomes from the serum of prostate cancer patients, which showed a significant increase in the levels of EpCAM and prostate-specific membrane antigen compared with healthy controls.
Size-based characterization of exosomes has also been intensely pursued. Compared with normal exosomes, cancer-exosome popu-lations show irregular morphologies and increased vesicle size124. Nanoparticle tracking analysis, which defines the particle size on the basis of Brownian motion, is the method most commonly used to determine the size distribution and concentration of isolated exosomes in suspension125. Another approach to separate exosomes on the basis of size uses deterministic lateral displacement through pillar arrays (Fig. 7d)126. Optimizing the silicon fabrication pro-cess allowed the production of nanoscale arrays with uniform gap sizes ranging from 25 to 235 nm, suitable for exosome sorting at sharp resolution.
Circulating tumour DNA fragments mainly originate from apop-totic tumour cells, which release their fragmented DNA into the blood circulation115,127–129. Elevated concentrations of ctDNA fragments have been found in the blood plasma and serum of cancer patients, and are being exploited for cancer screening130. However, control individuals also have mutated circulating DNA131. A recent study reported the observation of tumour protein 53-mutated cell-free