Open-Top Patterned Hydrogel-Laden 3D Glioma Cell Cultures for Creation of Dynamic Chemotactic Gradients to Direct Cell Migration
Aditya Rane, Steven TateJenna L. Sumey, Qing Zhong, Hui Zong, Benjamin Purow, Steven R. Caliari, Nathan S. Swami
The laminar flow profiles in microfluidic systems coupled to rapid diffusion at flow streamlines have been widely utilized to create well-controlled chemical gradients in cell cultures for spatially directing cell migration. However, within hydrogel-based closed microfluidic systems of limited depth (≤0.1 mm), the biomechanical cues for the cell culture are dominated by cell interactions with channel surfaces rather than with the hydrogel microenvironment. Also, leaching of poly(dimethylsiloxane) (PDMS) constituents in closed systems and the adsorption of small molecules to PDMS alter chemotactic profiles. To address these limitations, we present the patterning and integration of a PDMS-free open fluidic system, wherein the cell-laden hydrogel directly adjoins longitudinal channels that are designed to create chemotactic gradients across the 3D culture width, while maintaining uniformity across its ∼1 mm depth to enhance cell–biomaterial interactions. This hydrogel-based open fluidic system is assessed for its ability to direct migration of U87 glioma cells using a hybrid hydrogel that includes hyaluronic acid (HA) to mimic the brain tumor microenvironment and gelatin methacrylate (GelMA) to offer the adhesion motifs for promoting cell migration. Chemotactic gradients to induce cell migration across the hydrogel width are assessed using the chemokine CXCL12, and its inhibition by AMD3100 is validated. This open-top hydrogel-based fluidic system to deliver chemoattractant cues over square-centimeter-scale areas and millimeter-scale depths can potentially serve as a robust screening platform to assess emerging glioma models and chemotherapeutic agents to eradicate them.
Glioblastoma (GBM) is the most common and aggressive type of primary brain cancer in adults (1) that remains incurable and recurs frequently, (2) highlighting the need for in vitro patient-specific models that can predict disease outcomes. Recent work has correlated the infiltrative nature of the disease to glioma cell migration characteristics, (3) but recapitulation of the chemical and biomechanical cues that affect cell migration in the tumor microenvironment (TME) remains a major challenge within these models. (4) Based on hyaluronan, which is a primary extracellular matrix component in the brain, hyaluronic acid hydrogel networks are known to induce dose-dependent alterations to markers of glioma malignancy. (5) The inclusion of hyaluronic acid-based hydrogel as a matrix, with soluble CXCL12 as a chemoattractant for CXCR4-expressing GBM cells (6) and with chemotherapeutic agents to eradicate them, (7) is being investigated as a less invasive and more targeted pathway to remove residual glioma cells from the TME. To complement prior work on coupling laminar flow profiles to rapid diffusion at flow streamlines in closed microfluidic systems, (8) we present an open-top microfluidic platform to create chemotactic gradients for spatiotemporal control of glioma cell migration in a patterned hydrogel of millimeter-scale depth that is designed to enhance cell–biomaterial interactions.
Pressure-based microfluidic flow control in closed microchannels for cell culture within poly(dimethylsiloxane) (PDMS) backing layers on one side to enable fluidic access and bonded to a glass coverslip to enable access to live-cell imaging, is commonly used to investigate cellular processes. However, to create 3D cultures with biomechanical cues from the hydrogel-based cell microenvironment, without being limited by cell–cell interactions or cell–surface interactions, the cultures must be patterned over several cell layers (∼mm-scale depths) between the flows that deliver the chemotactic cues. The chief challenge to maintaining such 3D cultures in closed microfluidic systems over several days for directing cell migration (9) is the leaching of PDMS monomers into the culture medium, which affects cell proliferation, cell adhesion, and differentiation of cells in pharmacokinetic studies. (10−12) Furthermore, the adsorption of small hydrophobic molecules by PDMS over the long term of the cell culture affects their transport to the culture, thereby altering dose response and drug gradients. (13,14) Also, alterations in oxygen permeability with PDMS bonding (15) and its high permeability to water vapor (16,17) can cause culture media evaporation, drying, and bubble formation that are detrimental to the establishment of chemotactic gradients. (18) Alternate substrates for 3D cell culture, such as PMMA, COC, and adhesive tapes, usually involve cumbersome fabrication and assembly steps that limit rapid prototyping of device designs and assays. (19,20) The availability of open-top hydrogel-laden 3D cultures devoid of PDMS interfaces would mitigate many of these limitations, but current reports (21) do not yet integrate fluidic control operations for dynamic modulation of gradients that provide chemotactic cues to direct cell migration.
The patterning of cell-laden hydrogels for 3D culture in closed microfluidic systems is usually accomplished with arrays of microposts or pillars that use the surface tension of the hydrogel material to confine it between the adjoining media channels. To prevent the hydrogel from spilling into the adjoining fluidic channel, the injection pressure must be carefully controlled, (22−24) which depends on the viscosity, wettability, and other material properties of the hydrogel. High-aspect-ratio microfabrication is needed to create posts that extend several micrometers in the lateral scale and up to millimeters in the depth scale to contain cell-laden hydrogels for 3D culture. This can lead to discontinuities in the interface between the hydrogel and the perfused medium, thereby subjecting the cells to altered biochemical cues. (22) Recent approaches to create continuous interfaces of the hydrogel to the adjoining fluid in closed channels have emerged, such as the use of phase guides, stepped height channels, recoverable elastic barriers, and alignment of core–shells, but these require multilayer fabrication and assembly. (23−27) Rail-based capillary-pinning approaches for the patterning of open-top 3D cell cultures have been reported (21,28,29) but are static and without the fluidic control needed to deliver chemical gradients for dose/drug response assays or directed migration studies. Microfluidic probes (MFPs) to deliver gradients to open-top cell cultures by using injection and aspiration flows (30−38) require careful optimization of the geometry and flow rates of the probes. (35−40) Also, these are impractical for use in patterned 3D hydrogel cultures due to limited depth control of the confined fluid and depletion of the medium that immerses the 3D culture. To address these issues, we present a PDMS-free open microfluidic system (Figure 1A) integrating the patterned cell-laden hydrogel (∼1 mm depth) with adjoining longitudinal microchannels for dynamic flow control to create chemotactic gradients across the 3D culture width to direct glioma cell migration.
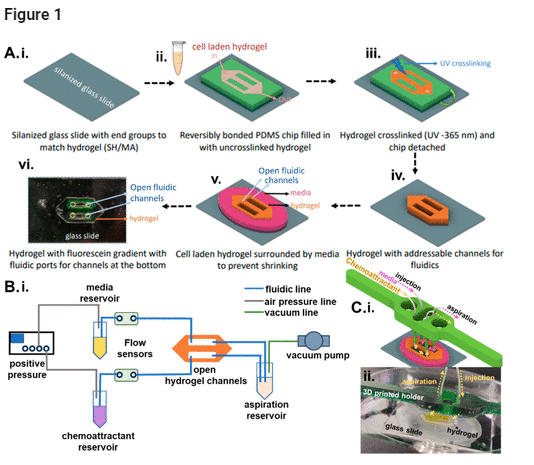
Figure 1. (A) Patterned cell-laden hydrogel adjoining fluidic channels. (i) A silanized glass substrate treated for adhesion to the cross-linked hydrogel is (ii) reversibly bonded to a PDMS mold that is then filled with the cell-laden hydrogel and (iii) photo-cross-linked to create the patterned hydrogel on glass. (iv) The PDMS mold is removed to leave open fluidic channels that directly adjoin the patterned hydrogel. (v) The structure is surrounded with culture medium to maintain cell viability and prevent hydrogel shrinkage. (vi) An example of the patterned hydrogel with addressable open fluidic channels through which a FITC gradient was established. (B) (i) Microfluidic flow control setup. (C) (i) 3D-printed holder for fluidic integration with the patterned hydrogel and (ii) image of the patterned hydrogel with tubing to the 3D-printed holder and channel with yellow dye.
While a reversibly bonded PDMS mold to glass is used as a receptacle to fill in and cure the hydrogel to enable its patterning, this PDMS layer is carefully detached after hydrogel curing, leaving behind only the patterned hydrogel layer adjoining the addressable fluidic channels. Another essential feature of our design is the integration of flow control (Figure 1B) over centimeter-scale lengths in the longitudinal open fluidic channels to deliver the culture medium and remove the waste products, while enabling dynamic modulation of the gradient of chemotactic molecules across the cell-laden hydrogel. This is accomplished through injection flow at one end and aspiration flow on the other end, which are placed in a 3D-printed construct to adjust the height of the microfluidic probes at the injection side to be below the hydrogel height level (Figure 1C(i,ii)), and the aspiration tubing to be just above the hydrogel height level, with surface tension confining the culture medium within the channels over the flow length. Rather than perfusing the culture with a peristaltic pump that creates pulsatile flow, which is difficult to monitor and control over the 48 h culture period, a pressure pump is used to deliver injection fluid at a continuous flow rate, and a vacuum line is used for the aspiration flow. In this manner, a set of integrated flow sensors can continually monitor any alterations and correct them by modulating the injection or aspiration flow rates, thereby maintaining an open cell culture within an incubator jacket, without drying of the hydrogel. Using a flow rate of 7.5 μL/min, which is close to the level reported for interstitial flow in brain tissues, (41−43) we present the ability to tune chemical gradients to the cells within the patterned hydrogel culture. This open-top system is validated using a patterned culture of U87 glioma cells laden within a hyaluronic acid (HA)–gelatin methacrylate (GelMA) hybrid hydrogel that is optimized to maintain cell viability, while including the adhesive groups necessary for integrin-mediated cell migration. A chemotactic gradient of CXCL12 delivered to the open-top cell-laden hydrogel culture is used to assay migration cues in the presence and absence of AMD3100, an inhibitor to CXCR4-expressing GBM cells. We envision utilization of this open-top integrated system to deliver chemotactic gradients and serve as a drug testing platform for micropatterned cell-laden hydrogel models.
Results and Discussion
Optimizing the Hydrogel Composition for Maintaining Cell Viability and Adhesion Motifs for Migration
Hyaluronan is a primary extracellular matrix component in the brain, leading to the interest in utilization of HA hydrogels to mimic the glioma microenvironment. However, while it can interact with cells through cell surface markers such as CD44, it lacks the adhesion motifs necessary for integrin-mediated cell migration. (44) Hence, we optimized a hybrid photo-cross-linked hydrogel composed of GelMA, which includes adhesive groups such as RGD, with HA to mimic the brain TME. As shown in Figure 2A, this is accomplished using lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) as a common photoinitiator for free-radical-initiated cross-linking of norbornene-modified HA (NorHA) and GelMA hydrogels. While GelMA hydrogels have higher stiffness, (45) HA hydrogels have been used to mimic the stiffness of native brain tissue. (46) As shown in Figure 2B, this photopatterned hybrid hydrogel, consisting of 2% GelMA and 1% NorHA, exhibits a Young’s modulus that mimics the brain’s white matter and gray matter components. (47) Based on propidium iodide staining (Figure 2C), GelMA hydrogels (10%) support U87 cells to a viability level of only 60% over the 48 h culture period, whereas the hybrid hydrogel supports ∼75% cell viability, which is closer to the 90% viability levels observed within 1% NorHA hydrogels.
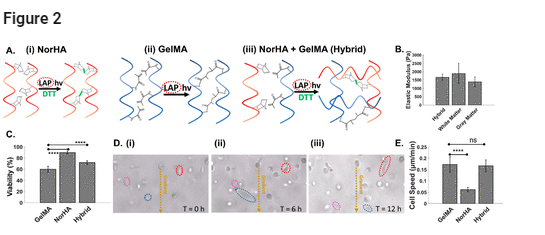
Figure 2. (A) Individual and hybrid hydrogels. Molecular structure before and after cross-linking of (i) norbornene-modified hyaluronic acid (norHA), (ii) gelatin methacrylate (GelMA), and (iii) a hybrid hydrogel of NorHA and GelMA. (B) Young’s modulus of the hybrid hydrogel formulation (2% HA, 1% GelMA) compared to that of brain tissue. The hybrid hydrogel recapitulates the reported stiffness of white and gray matter. (C) Cell viability as a function of hydrogel composition. (D) Cell mobility based on cell migration through the hydrogel at the indicated time points (i)–(iii). (E) Measured cell migration speed as a function of hydrogel composition. Significance was determined by one-way ANOVA with Tukey’s post-hoc test, represented as ns, p ≥ 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
Differences in pore size between the respective hydrogels can affect cell viability, with 1% NorHA hydrogels reported at a theoretical mesh size of ∼85 nm, (46) while mesh sizes of 10% GelMA are reported as ∼20 nm. (48) However, this difference in mesh size, as reported for fully swollen hydrogels, is likely less pronounced in our study, since the hydrogels are confined in the device channels and are somewhat restricted from swelling. Instead, GelMA hydrogels that are formed by chain-growth cross-linking are inhibited by oxygen and require a high concentration of free radicals for initiation of polymerization, (49) leading to a loss in viability. On the other hand, step-growth-polymerized NorHA hydrogels require lower free radical concentrations and are not inhibited by oxygen, (50) thereby resulting in faster cross-linking and improved cytocompatibility. (51) This hybrid hydrogel also supports viability of a 3D culture of highly migratory and malignant oligodendrocyte progenitor cells (OPCs) that are progenitors in glioma (52,53) (Figure S1). Based on time-lapse images of U87 cell alignment and migration (Figure 2D(i–iii)), the cell speeds in the hybrid hydrogel resemble those observed within the GelMA hydrogel, rather than the HA hydrogel (Figure 2D(iv) and Movie S1). This indicates the optimization of the hybrid hydrogel for its high viability and migration abilities.
Open-Top Microfluidics for Spatiotemporal Control of Chemotactic Gradients Across Patterned Hydrogel
The open-top 3D culture was utilized with longitudinal injection and aspiration flows to deliver and control chemotactic gradients across the patterned hydrogel width. Using fluorescein isothiocyanate (FITC)-labeled dextran (FITC-dextran) with a 400 Da molecular weight to mimic small-molecule drugs, the images (Figure 3A(i,ii)) show development of the gradient across the hydrogel width. Using FITC-dextran with a 10 kDa molecular weight to mimic proteins or cytokines, fluorescence levels measured from time-lapse images show molecular diffusion profiles at different widths across the patterned hydrogel, indicating the ability of the open-top microfluidic system to induce gradual flattening of the initial diffusion profile. It is apparent that the 10 kDa FITC-dextran takes well over 12 h to reach a steady-state profile (Figure 3B(ii)), whereas the 400 Da FITC-dextran reaches deep into the hydrogel within an hour (Figure 3B(i)). This chemotactic gradient can be created across the complete depth of a 1 mm thick hydrogel, as apparent from similar Z-stack FITC levels (Figures 3C and S2 and Movie S2) at the top and bottom of the hydrogel (normalized to FITC level at the center). Also, this gradient can be sustained across a large width of the hydrogel (∼2 mm), with an ∼15 mm long uninterrupted and continuous interface between the fluid and hydrogel, whereas prior work had used posts. Hence, the open microfluidic system can create well-defined chemical gradients to viable glioma cells over the long durations needed to assay cell responses (48 h).
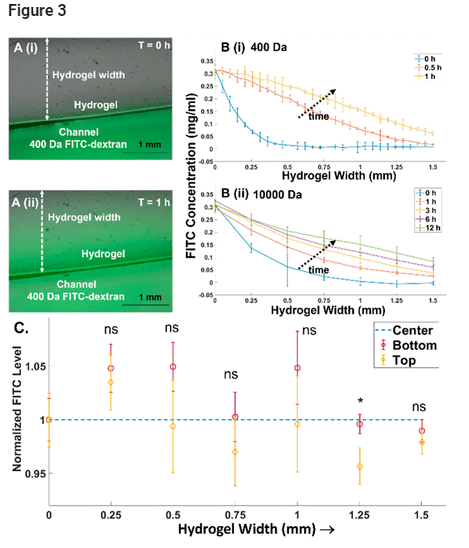
Figure 3. (A) Bright-field and FITC overlay of the gradient across the hydrogel at various time points with 400 Da FITC-dextran at (i) T = 0 h and (ii) T = 1 h. (B) Temporal development of the gradient across the open hydrogel for (i) 400 Da FITC-dextran, with the gradient developing rapidly to approach steady state within 1 h, and (ii) 10000 Da FITC-dextran, with the gradient developing slowly over 12 h. (C) The chemical gradient develops over the entire hydrogel depth (1 mm) based on the similar FITC levels at the top, bottom, and center of the hydrogel, and its invariance over the hydrogel width at steady state (24 h) for 10000 Da FITC-dextran. Significance is based on one-way ANOVA with Tukey’s post-hoc test, represented as ns, p ≥ 0.05; *, p ≤ 0.05.
Migration Cues to Glioma Cells in the Patterned Hydrogel Using Open-Top Microfluidics
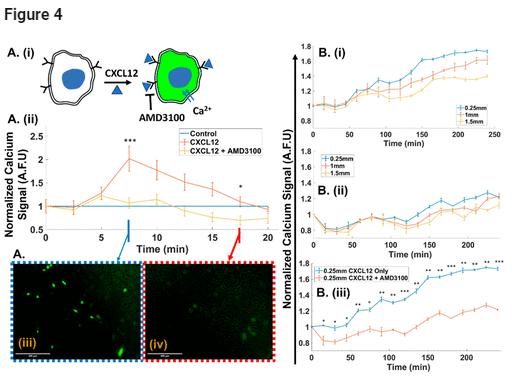
To assess the ability of the open-top microfluidic system to deliver chemotactic gradients for cues to cells across the hydrogel width, we utilized the chemokine: CXCL12, which induces migration of CXCR4-expressing glioma cells, and AMD3100, which inhibits this signaling pathway. (54) As shown in the schematic in Figure 4A(i), gradients of CXCL12 induce calcium influx into the cell upon binding to its cell membrane receptor. Hence, glioma cells labeled with a calcium signaling probe that fluoresces upon calcium binding were used to quantify effect of the chemotactic gradient on cells across the hydrogel width. Fluorescence image analysis of a static culture of glioma cells was used to determine the CXCL12 level in the medium that is needed for signal rise above the baseline. Similarly, inhibition of CXCL12 stimulation at this level was validated using cells pretreated with 1 μg/mL AMD3100. (54) Based on fluorescence signal plots (Figure 4A(ii)) and images (Figure 4A(iii,iv)), we infer that 66 ng/mL CXCL12 in the medium is sufficient to cause signal rise within 5 min of stimulation, and this signal rise is effectively inhibited for cells pretreated with 1 μg/mL AMD3100. Beyond a critical time in the static culture, there is a gradual signal dropoff to the baseline level, similar to that under no CXCL12 stimulation. This is attributed to CXCL12 diffusional limitations, since this is not apparent in the dynamic 3D culture that constantly is replenished with CXCL12. When 66 ng/mL CXCL12 is used in the medium under dynamic 3D culture (7.5 μL/min perfusion) to create a gradient across the width of the cell-laden hydrogel, the fluorescence signal (Figures 4B(i) and S3) develops rapidly at proximal channel widths (0.25 mm) while taking longer to extend to 1 and 1.5 mm widths. With cells pretreated with 1 μg/mL AMD3100, the fluorescence level is diminished at the same widths (Figure 4B(ii)). Comparison of the normalized fluorescence signal with CXCL12 shows a 1.8-fold increase over the control, while the level remains close to the control after inhibitor pretreatment (Figure 4B(iii)). This validates ability of the open-top microfluidic system to deliver well-controlled cues from the CXC12 gradient to cells across the hydrogel width and cause its inhibition with AMD3100.